| Location within campus: | Unidad de Morfología y Función |
| Phone: | 55 56 23 12 60 y 55 59 64 76 80 |
| Head of Laboratory: | Dr. Roberto Velasco García Full Professor robertvela2001@yahoo.com.mx |
| Researchers affiliated to the laboratory: | M. Sc. María del Rocío Vargas Martínez Associate Professor procionida@yahoo.com.mx |
| Technicians affiliated to the laboratory: | Dr. Edaena Benítez Rangel edabenitez@gmail.com |
| Research lines per researcher: | Biochemical-structural characterization of the glucose-6-phosphate dehydrogenase enzyme from Pseudomonas aeruginosa. Structural and functional characterization of the HexR transcriptional repressor of Pseudomonas aeruginosa. |

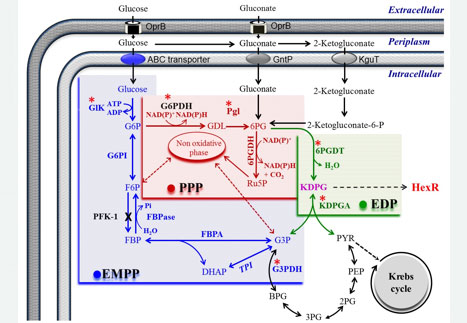
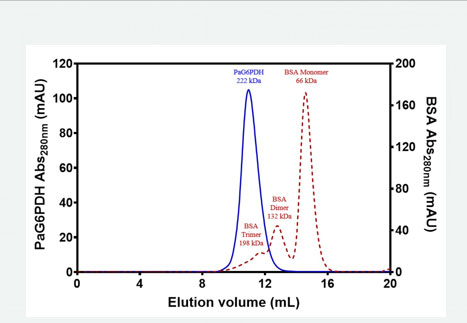
In the Osmoregulation Laboratory of the FES Iztacala we studied the glucose-6-phosphate dehydrogenase of Pseudomonas aeruginosa (G6PDHPa), an enzyme that produces NADPH that this microorganism requires to face oxidative stress when it invades humans. In the search for substances that would help control the infections caused by the pathogen, the role that this enzyme plays has led us to consider it a potential target. In addition to exploring possible inhibitors of its activity, we also investigate substances and conditions that could affect its structural stability.
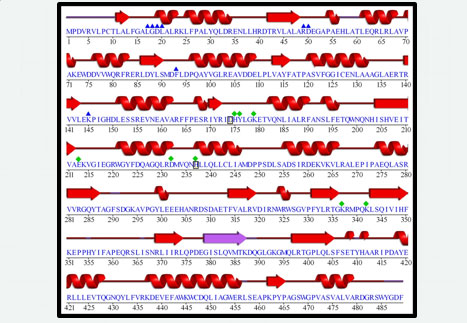
Other points of G6PDHPa that we address have to do with the elucidation of its three-dimensional structure and the knowledge of amino acid residues that could be important for its structure and its function.
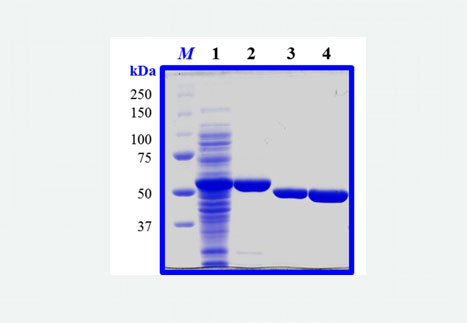
In another investigation parallel to the G6PDHPa study, we recently cloned the P. aeruginosa hexR gene, and overexpressed and purified the recombinant HexRPa protein from E. coli. It has been proposed, for other bacterial species, that this protein transcriptionally regulates the expression of zwf, which codes for G6PDHPa, and that of other genes found in the same operon and that code for enzymes that participate in the same metabolic cycle as G6PDHPa , called EDEMP. We will shortly begin the structural and functional characterization of HexRPa to find out if it plays the same role in P. aeruginosa as in other species.

Velasco-García, R. and Vargas-Martínez, R. (2012). The study of protein–protein interactions in bacteria. Can. J. Microbiol., 58, 1241–1257. https://doi.org/10.1139/w2012-104
Kevin E. Acero-Navarro, K. E., Jiménez-Ramírez, M., Villalobos, M. A., Vargas-Martínez, R., Perales-Vela, H. V. and Velasco-García, R. (2018). Cloning, overexpression, and purification of glucose-6-phosphate dehydrogenase of Pseudomonas aeruginosa. Protein Expression and Purification, 142, 53-61.
https://doi.org/10.1016/j.pep.2017.10.004
Garrido, F., Pacheco, M., Vargas-Martínez, R., Velasco-García, R., Jorge, I., Serrano, H., Portillo, F., Vázquez, J. and Pajares M. A. (2018). Identification of hepatic protein-protein interaction targets for betaine homocysteine S-methyltransferase. Plos One, 13(6), e0199472.
https://doi.org/10.1371/journal.pone.0199472
Benítez-Rangel, E., Rodríguez-Hernández, A. and Velasco-García, R. (2020). The substrate of the glucose-6-phosphate dehydrogenase of Pseudomonas aeruginosa provides structural stability. BBA-Proteins and Proteomics 1868, 140331.
https://doi.org/10.1016/j.bbapap.2019.140331

| Entry profile of potential thesis students: | Students who intend to enter the Osmoregulation Laboratory as thesis must comply, preferably, with the following: • Be interested in studying the structure and function of proteins and enzymes • Be a regular student (do not owe courses) • Have a minimum average of eight (8.0) • Take the subjects LICyT VII and LICyT VIII in the Osmoregulation laboratory, in order to carry out thesis work during the 7th and 8th semesters. |
| Service Social Data: | “ACTIVITIES TO SUPPORT THE STRUCTURAL BIOCHEMICAL STUDY OF THE ENZYME GLUCOSE-6-PHOSPHATE DEHYDROGENASE (G6PDH) OF PSEUDOMONAS AERUGINOSA AND ITS INTERACTION WITH EFFECTORS AND INHIBITORS OF ITS ACTIVITY”. KEY 2020-12 / 63-33 ACTIVITIES: • Preparation of media for bacteria culture. • Obtaining enzymatic variants by site-specific mutagenesis. • Culture of transforming bacteria and overexpression of recombinant proteins. • Purification and conservation of recombinant enzymes. |